How can targeting proteins, not only genes, revolutionize disease treatment?

Cancer has long been perceived as a genetic disease at large –that is, cancer initiation, promotion and progression is dictated by certain genetic changes (mutations) [1]. Numerous studies [2] have identified nearly 1000 cancer-associated genes so far (~250 oncogenes and ~700 tumor suppressor genes). Knowing that a cell needs at least two mutations in such genes to become cancerous, more than 1 million cancer genotypes would be generated. How can we treat millions of cancer genotypes with few therapeutics in hand, even fewer aimed at such genes – genes that are ever-mutating?
An estimate of only 6.4% of patients were successfully paired with mutation-targeted therapeutics following genetic analysis, according to one study [3]. Another study [4] revealed that tailoring the treatment of advanced gastric cancer according to patients’ tumor genetics extended survival by only 2.9 months.
New studies [5] demonstrate that regardless of the heterogeneous, cancer-triggering genetic and epigenetic alterations, the “program of gene expression” –and related protein activity that sustains a tumor for a given cancer type – is almost identical from one patient to another, even when compared with respective normal tissue. “Mass regulator” (MR)[6] proteins are considered part of that conserved program. After all, mutations are somewhat static, while proteins’ production is dynamic and, therefore, represents a real-time window into cancer activity. Such studies usher in a new and promising era of precision medicine that targets proteins, not only mutations, to impair cancer growth, if not kill off cancer cells.
Proteins are not only emerging as a valuable target in cancer therapy, but also as an effective tool to aid disease diagnosis, predict response to treatment, and mitigate treatment toxicity. Technology has come far to allow for hundreds of proteins to be screened simultaneously. Antibody-based assays such as multiplex immunoassays (ELISA) can provide simultaneous quantitative measurement of various proteins (cytokines, growth factors, proteases, soluble receptors, etc.) in a wide variety of sample types.
Proteomic signatures to diagnose disease
Moledina and colleagues [7] found that patients with acute interstitial nephritis (AIN) (i.e., an allergic reaction to medication in the kidney) have in their urine – not blood – interleukin (IL)-9, which is produced by the mast cells (immune cells), and instigates an allergic response by releasing histamine. This has the potential to evolve into a clinical urine test for AIN diagnosis, if validated in larger studies. The standard method to diagnose AIN is through kidney biopsy, which is an invasive procedure and can be associated with complications. Furthermore, AIN, alongside other kidney diseases, can be hard to diagnose given the fact that if notable portions of the kidney were damaged, the patient would not experience obvious symptoms, as the presence of two kidneys allows for the second kidney to compensate for the first, damaged one.
Similar eye symptoms can arise from four different diseases (metastatic lymphoma, bacterial, fungal, or viral infection), which cannot be differentiated through clinical imaging. However, a protein assay of the vitreous (eye) fluid can show different proteomic signatures [8-10], which can distinguish each disease apart from the other; allowing physicians to arrive at a diagnosis and treatment plan much faster. As one example, early-stage proliferative vitreoretinopathy (PVR) can be set apart from late-stage PVR. Early-stage was shown to be associated with T-cell markers, cytokines downstream of mTOR activation (IL2, 6, and 13), and pro-fibrotic cytokines. Whereas, late-stage was associated with cytokines driving monocyte responses and stem-cell recruitment (SDF-1) [8].
Proteomic biomarkers to predict response to treatment
Homicsko and colleagues [11] used an antibody array to test the serum of patients with advanced metastatic melanoma for 440 various proteins. The patients were given immunotherapy ((Yervoy®) ipilimumab: a CTLA-4 inhibitor, and (Opdivo®) nivolumab: a PD-1 inhibitor), and their progress was followed for 36 months. The analysis showed three protein biomarkers (FGF4, CCL15, and TACE) that were correlated with responses to cancer immunotherapy. In 90% of patients possessing these protein biomarkers, the treatment halted the cancer for at least 3 years, compared with patients lacking these protein biomarkers.
Proteomic markers to mitigate treatment toxicity
When cancer patients are treated with chemotherapy or radiotherapy, their bone marrow stem cells suffer. Himburg and colleagues [12] found that a protein termed Dkk1 was able to protect the bone marrow against radiation damage in mice by speeding the recovery of blood cells and lowering the incidence of infection. Their studies also showed that Dkk1 acts directly on stem cells by suppressing their death, and indirectly by stimulating endothelial cells to generate epidermal growth factor, which supports stem cell regeneration. When they gave lethally-irradiated mice Dkk1, their blood cells’ recovery was enhanced.
Mass regulator (MR) proteins to treat cancer
By analyzing the gene expression profiles of cells from tumor samples and applying information theory mathematics to the emerging data, MRs were discovered [6]. Califano and colleagues analyzed data from 20,000 tumor samples, generating maps for 36 tumor types, and identifying approximately 300 proteins that can be considered MRs in at least one type of cancer. MRs are mostly proteins that influence transcription. Califano and Alvarez [6] described MRs as a few proteins that control the largest numbers of other proteins and the stability of cancer states, independent of certain genetic mutations. Many of these MRs, once activated, never shut down, through establishing self-feedback loops. Examples include aggressive prostate cancer, in which two proteins, FOXM1 and CENPF, act synergistically to promote tumor growth through such feedback loops [13]; high grade glioblastoma (the most aggressive form of brain cancer), in which three proteins were shown to start and maintain cancer growth [14] (Fig. 1); and small-cell lung cancer, in which two MRs (POU2F3 and ASCL1) were identified [15].
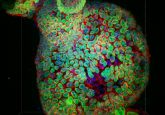
Array
Figure 1. Shows how very few proteins can control large protein networks independent of genetic mutations and, hence, dictate the stability of cancer states. In the mesenchymal signature of high-grade glioma, CCAAT/enhancer- binding protein (CEBP) controls 74% of the genes.
The discovery of MRs marks an uncharted territory in cancer treatment – one that is patient-personalized, and if targeted effectively, has the potential to extend or save many more lives. Their discovery also chips away at the limited perspective that cancer is a genetic disease. Be that as it may, MRs can be in classes of proteins that are difficult to therapeutically-target, and multiple MRs might need to be targeted simultaneously to have a tumor regression effect. Further, MRs might act in animal models differently than in humans. Several studies are underway to address these concerns.
Current technologies facilitating the screening and quantification of hundreds of proteins allow them to take center stage in precision and personalized medicine whereby proteins play a key role in enhancing disease diagnosis and cancer treatment, aiding the prediction of response to immunotherapy, and alleviating toxicity of chemotherapy and radiotherapy.
Suggested reading:
Califano A, Alvarez MJ. The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat. Rev. Cancer 17(2), 116–130 (2017). Link: www.nature.com/articles/nrc.2016.124
References:
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 9(4), 138–141 (1993).
- Forbes SA, Beare D, Gunasekaran P et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43(Database issue), D805–D811 (2015).
- Prasad V. Perspective: The precision-oncology illusion. Nature 537(7619), S63 (2016).
- Lee J, Kim ST, Kim K et al. Tumor genomic profiling guides metastatic gastric cancer patients to targeted treatment: The VIKTORY Umbrella Trial. Cancer Discov. 9(10), 1388–1405 (2019).
- Hoadley KA, Yau C, Wolf DM et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158(4), 929–944 (2014).
- Califano A, Alvarez MJ. The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat. Rev. Cancer 17(2), 116–130 (2017).
- Moledina DG, Wilson FP, Pober JS et al. Urine TNF-alpha and IL-9 for clinical diagnosis of acute interstitial nephritis. JCI Insight 4(10), 127456 (2019).
- Roybal CN, Velez G, Toral MA et al.Personalized proteomics in proliferative vitreoretinopathy implicate hematopoietic cell recruitment and mTOR as a therapeutic target. Am. J. Ophthalmol. 186, 152–163 (2018).
- Velez G, Yang J, Li AS et al. Proteomic insight into the pathogenesis of CAPN5-vitreoretinopathy. Scientific Reports-Uk 9, 7608 (2019).
- Mahajan VB, Skeie JM. Translational vitreous proteomics. Proteom. Clin. Appl. 8(3–4), 204–208 (2014).
- Homicsko K, Cuendet MA, Mlynska A et al. Exploratory analysis of multiprotein serum predictors at baseline of progression-free survival of ipilimumab or ipilimumab and nivolumab in the Checkmate-069 study. J. Clin. Oncol. 35(15), 9571–9571 (2017).
- Himburg HA, Doan PL, Quarmyne M et al. Dickkopf-1 promotes hematopoietic regeneration via direct and niche-mediated mechanisms. Nat. Med. 23(1), 91–99 (2017).
- Aytes A, Mitrofanova A, Lefebvre C et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell 25(5), 638–651 (2014).
- Tome-Garcia J, Erfani P, Nudelman G et al. Analysis of chromatin accessibility uncovers TEAD1 as a regulator of migration in human glioblastoma. Nat. Commun. 9 (1), 4020 (2018).
- Huang YH, Klingbeil O, He XY et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Gene Dev. 32(13–14), 915–928 (2018).
Author profile: Balkees Abderrahman, M.D. is the Dallas/Ft. Worth Living Legend Fellow of Cancer Research at the Department of Breast Medical Oncology, the University of Texas MD Anderson Cancer Center (TX, USA) and a PhD trainee under model “Individuals of Very High Quality” at the Faculty of Biological Sciences, the University of Leeds (West Yorkshire, UK). Balkees is a regular contributor to Oncology Central; you can view her full biography here.